library(PKPDsim)
library(clinPK)
library(dplyr)
library(tidyr)
library(ggplot2)
# remotes::install_github("mtennekes/cols4all")
library(cols4all)
cbpal <- c4a("kelly", 9) # color scheme for plots (colour-blind friendly)
# Make sure the following 9 models are installed, or adapt as appropriate.
models <- c(
"pkvancojacqzaigrain",
"pkvancokloprogge",
"pkvancocolin",
"pkvancocapparelli",
"pkvancofrymoyer",
"pkvancole",
"pkvancoanderson",
"pkvancodao",
"pkvancogermovsek"
)Visualizing clearance differences between neonate PK models with clinPK and PKPDsim
clinPK
PKPDsim
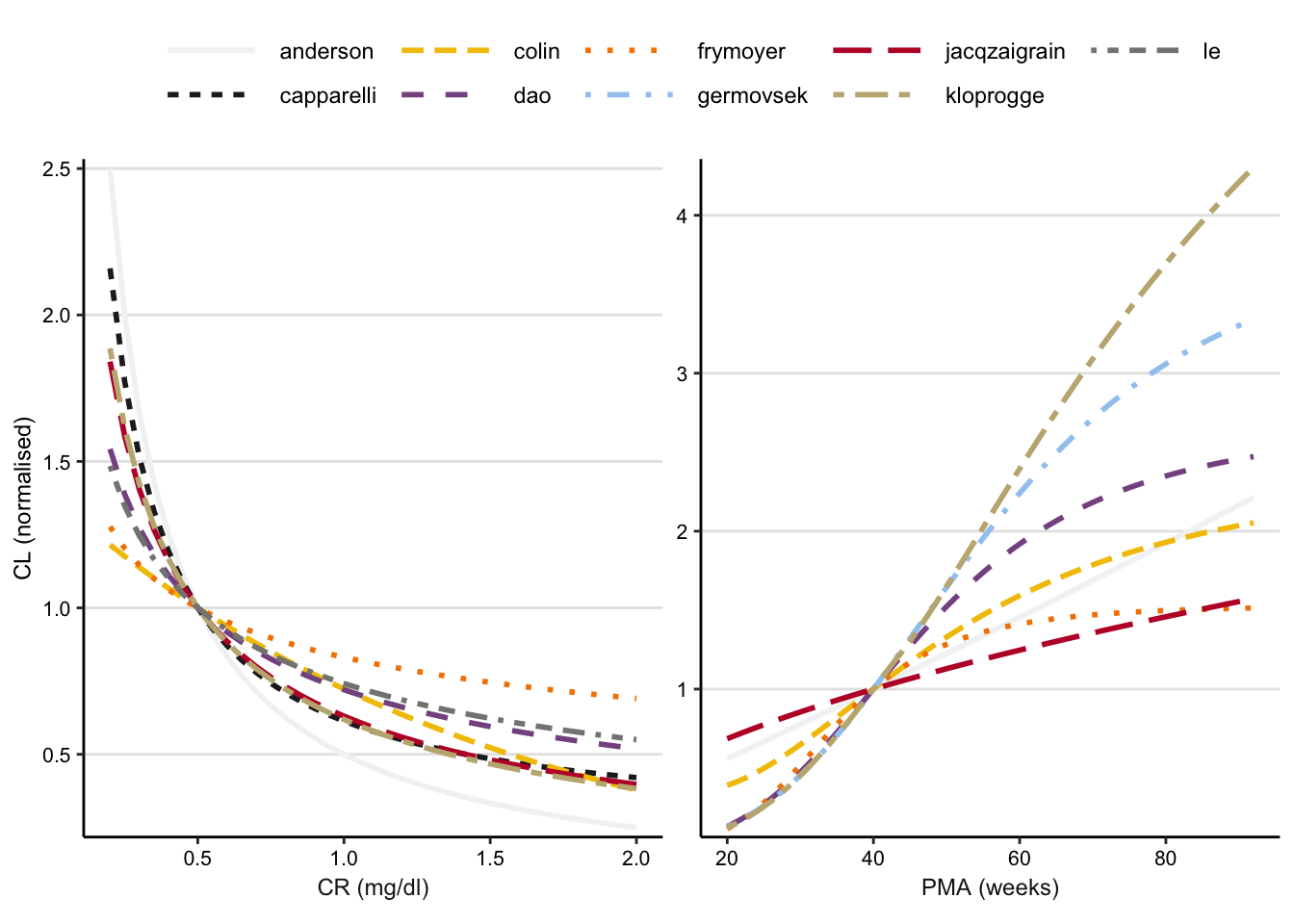
Calculate clearance for a given post-menstrual age (PMA) or serum creatinine, holding constant all other covariates, for 9 different PK models.
# ------------------------------------------------------------------------------
# Define functions
# ------------------------------------------------------------------------------
# Function to load the information we need for simulation for each model
# model_lib: name of model library, eg "pkvancokloprogge"
# what: objects to load from the model package namespace
load_model_objects <- function(
model_lib,
what = c("model", "parameters")
) {
suppressMessages( ## avoid message "the following objects are masked from ..."
require(model_lib, character.only = TRUE)
)
mod_objects <- list()
for(i in seq(what)) {
mod_objects[[what[i]]] <- get(what[i], asNamespace(model_lib))()
}
mod_objects
}
# Calculate CL for a particular CR
# model_lib: name of model library, eg "pkvancokloprogge"
# crs: vector of creatinine values for which to calculate CL
CL_by_CR <- function(model_lib, crs){
mod_obj <- load_model_objects(model_lib)
covs <- list(
SEX = new_covariate(1),
WT = new_covariate(10),
PMA = new_covariate(40),
GA = new_covariate(36),
AGE = new_covariate(4 * 7/365),
HT = new_covariate(70),
CL_HEMO = new_covariate(0),
CR_ASSAY = new_covariate(0)
)
vapply(
crs,
function(x) {
covx <- covs
crcl <- suppressMessages(suppressWarnings(clinPK::calc_egfr(
method = "schwartz_revised",
sex = "male",
weight = 10,
height = 70,
scr = x,
age = 0.2
)))
covx$CR <- new_covariate(x)
covx$CRCL <- new_covariate(crcl)
calculate_parameters(
mod_obj$model,
mod_obj$parameters,
covx
)[["CLi"]]
},
FUN.VALUE = numeric(1)
)
}
# Calculate CL for a particular PMA
# model_lib: name of model library, eg "pkvancokloprogge"
# pmas: vector of post-menstrual ages for which to calculate CL
CL_by_PMA <- function(model_lib, pmas){
mod_obj <- load_model_objects(model_lib)
modlib <- mod_obj$model_lib
crcl <- suppressWarnings(suppressMessages(
clinPK::calc_egfr(
method = "schwartz_revised",
sex = "male",
weight = 10,
height = 70,
scr = 0.7,
age = 0.2
)
))
covs <- list(
CR = new_covariate(0.7),
SEX = new_covariate(1),
WT = new_covariate(10),
HT = new_covariate(70),
CRCL = new_covariate(crcl),
CL_HEMO = new_covariate(0),
CR_ASSAY = new_covariate(0)
)
vapply(
pmas,
function(x) {
covx <- covs
covx$PMA <- new_covariate(x)
covx$GA <- new_covariate(pmin(x, 36))
covx$AGE <- new_covariate(pmax((covx$PMA$value - covx$GA$value) / 7, 1))
calculate_parameters(
mod_obj$model,
mod_obj$parameters,
covx
)[["CLi"]]
},
FUN.VALUE = numeric(1)
)
}# ------------------------------------------------------------------------------
# Run simulations
# ------------------------------------------------------------------------------
models_for_CR <- setdiff(models, "pkvancogermovsek")
models_for_PMA <- setdiff(models, c("pkvancole", "pkvancocapparelli"))
crs <- seq(0.2, 2, 0.05)
pmas <- seq(20, (40 + 52), 1)
# 1. Calculate CL at each value of covariate
cls_by_crs <- setNames(
lapply(
models_for_CR,
CL_by_CR,
crs
),
gsub("pkvanco", "", models_for_CR)
) %>%
as.data.frame() %>%
mutate(CR = crs)
cls_by_pmas <- setNames(
lapply(
models_for_PMA,
CL_by_PMA,
pmas
),
gsub("pkvanco", "", models_for_PMA)
) %>%
as.data.frame() %>%
mutate(PMA = pmas)
# 2. Normalize clearance to clearance at selected standard values
cls_cr05 <- cls_by_crs %>%
pivot_longer(-CR, names_to = "model", values_to = "CL") %>%
filter(CR == 0.5) %>%
select(model, cr05 = CL)
cls_by_crs_norm <- cls_by_crs %>%
pivot_longer(-CR, names_to = "model", values_to = "CL") %>%
left_join(cls_cr05, by = "model") %>%
group_by(model) %>%
mutate(CLn = CL/cr05) %>%
ungroup() %>%
mutate(model = gsub("_.*", "", model)) %>%
mutate(cov = "CR (mg/dl)") %>%
rename(value = CR)
cls_pma40 <- cls_by_pmas %>%
pivot_longer(-PMA, names_to = "model", values_to = "CL") %>%
filter(PMA == 40) %>%
select(model, pma40 = CL)
cls_by_pma_norm <- cls_by_pmas %>%
pivot_longer(-PMA, names_to = "model", values_to = "CL") %>%
left_join(cls_pma40, by = "model") %>%
group_by(model) %>%
mutate(CLn = CL/pma40) %>%
ungroup() %>%
mutate(model = gsub("_.*", "", model)) %>%
mutate(cov = "PMA (weeks)") %>%
select(model, cov, value = PMA, CLn)# ------------------------------------------------------------------------------
# Plot clearance as a function of covariate values
# ------------------------------------------------------------------------------
p <- bind_rows(cls_by_crs_norm, cls_by_pma_norm) %>%
ggplot() +
aes(x = value, y = CLn, color = model, linetype = model) +
facet_wrap(~cov, scales = "free", strip.position = "bottom") +
geom_line(size = 1) +
theme_classic() +
scale_color_manual(values = cbpal) +
ylab("CL (normalised)") +
theme(
legend.position = "top",
legend.title = element_blank(),
legend.key.width = unit(1.5, 'cm'),
panel.grid.major.y = element_line(colour = "grey90"),
axis.text = element_text(size = 8, color = 'black'),
axis.title = element_text(size = 9, color = 'black'),
axis.title.x = element_blank(),
plot.tag = element_text(size = 10, family = "Times"),
strip.text = element_text(size = 9),
strip.background = element_blank(),
strip.placement = "outside",
strip.text.x = element_text(margin=margin(b=5))
) +
scale_y_continuous(expand = c(0.01,0.01))
p